-
- Downloads
Revised top n genera figures for all markers
Revision request, reknitted as a result
Showing
- analysis/16S/16S_Analysis_pipeline_v4-3.Rmd 15 additions, 17 deletionsanalysis/16S/16S_Analysis_pipeline_v4-3.Rmd
- analysis/16S/16S_Analysis_pipeline_v4-3.html 125 additions, 142 deletionsanalysis/16S/16S_Analysis_pipeline_v4-3.html
- analysis/16S/16S_DADA2_results_260821/analysis_output/Controls.pdf 0 additions, 0 deletions...16S/16S_DADA2_results_260821/analysis_output/Controls.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Controls_13-03-25_160718.pdf 0 additions, 0 deletions...sults_260821/analysis_output/Controls_13-03-25_160718.pdf
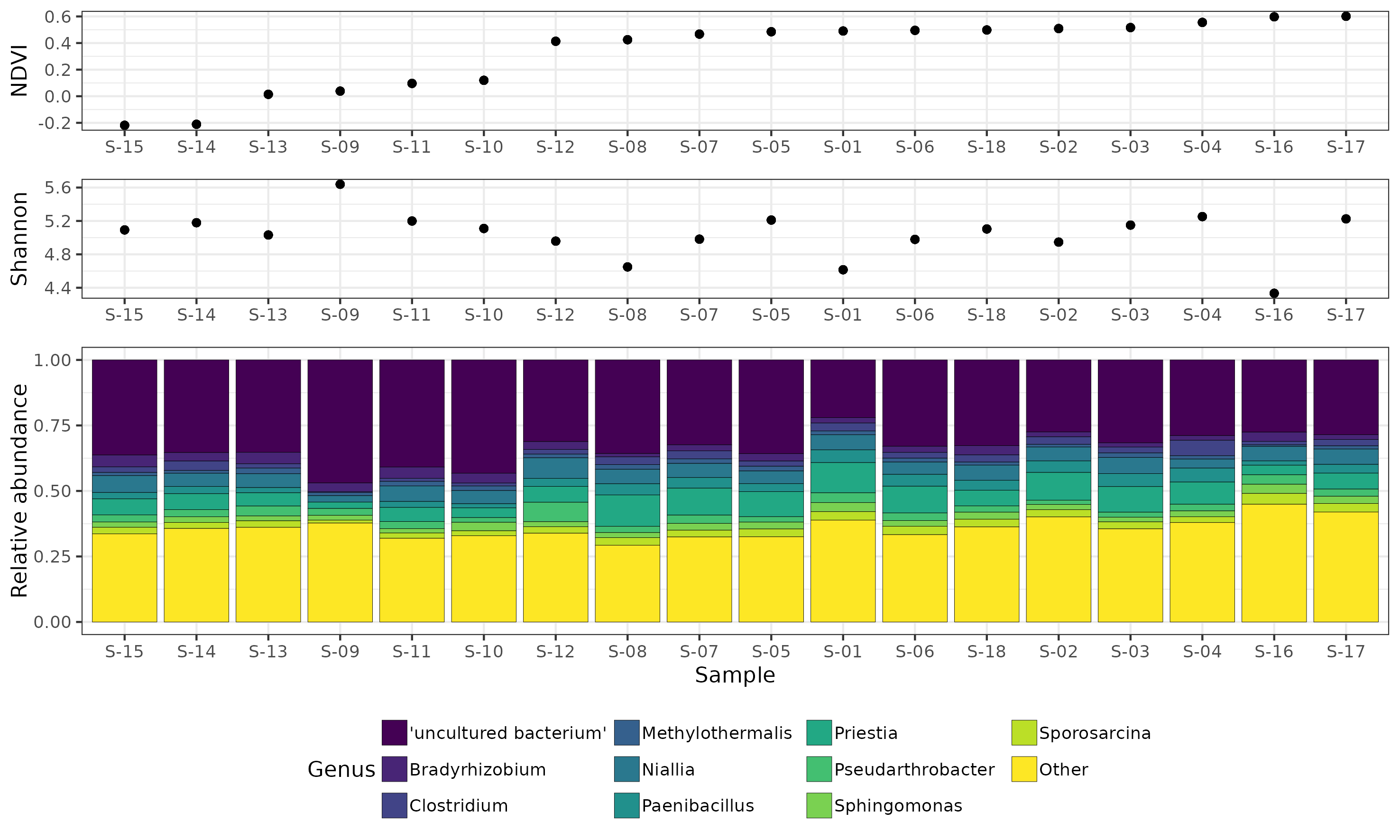
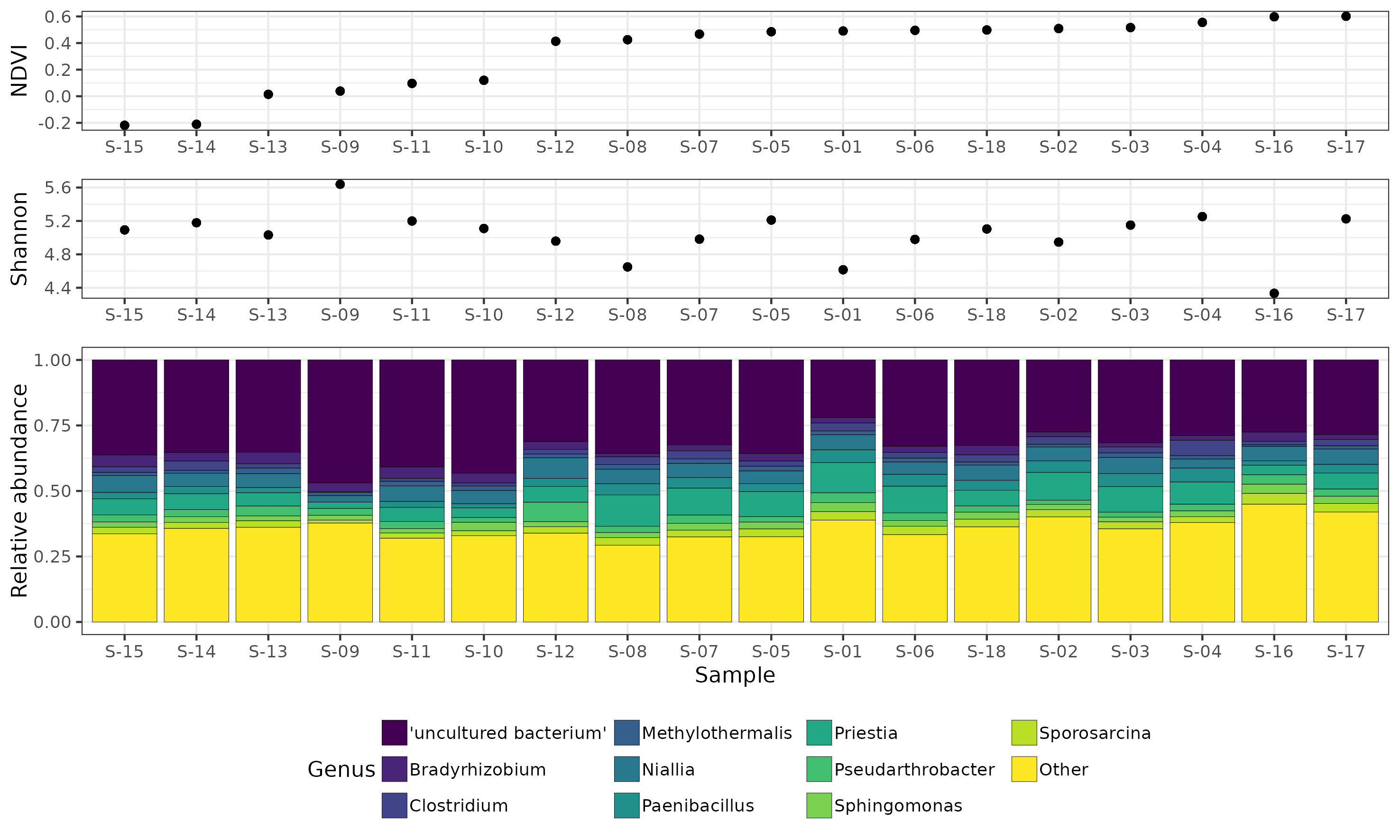
- analysis/16S/16S_DADA2_results_260821/analysis_output/Customized_NDVI_Shannon_plot.pdf 0 additions, 0 deletions...s_260821/analysis_output/Customized_NDVI_Shannon_plot.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Customized_NDVI_Shannon_plot.png 0 additions, 0 deletions...s_260821/analysis_output/Customized_NDVI_Shannon_plot.png
- analysis/16S/16S_DADA2_results_260821/analysis_output/Customized_NDVI_Shannon_plot_13-03-25_160939.pdf 0 additions, 0 deletions...s_output/Customized_NDVI_Shannon_plot_13-03-25_160939.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Customized_NDVI_Shannon_plot_13-03-25_162547.pdf 0 additions, 0 deletions...s_output/Customized_NDVI_Shannon_plot_13-03-25_162547.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Customized_NDVI_Shannon_plot_13-03-25_163029.pdf 0 additions, 0 deletions...s_output/Customized_NDVI_Shannon_plot_13-03-25_163029.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Customized_NDVI_Shannon_plot_17-03-25_101300.png 0 additions, 0 deletions...s_output/Customized_NDVI_Shannon_plot_17-03-25_101300.png
- analysis/16S/16S_DADA2_results_260821/analysis_output/Overview_all_and_pruned.pdf 0 additions, 0 deletions...esults_260821/analysis_output/Overview_all_and_pruned.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Overview_all_and_pruned_13-03-25_160716.pdf 0 additions, 0 deletions...alysis_output/Overview_all_and_pruned_13-03-25_160716.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Top100_ASVs.pdf 0 additions, 0 deletions.../16S_DADA2_results_260821/analysis_output/Top100_ASVs.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Top100_ASVs_13-03-25_160726.pdf 0 additions, 0 deletions...ts_260821/analysis_output/Top100_ASVs_13-03-25_160726.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Top100_Genus.pdf 0 additions, 0 deletions...16S_DADA2_results_260821/analysis_output/Top100_Genus.pdf
- analysis/16S/16S_DADA2_results_260821/analysis_output/Top100_Genus_13-03-25_160725.pdf 0 additions, 0 deletions...s_260821/analysis_output/Top100_Genus_13-03-25_160725.pdf
- analysis/16S/genus_abundance_tbl_per_sample.xlsx 0 additions, 0 deletionsanalysis/16S/genus_abundance_tbl_per_sample.xlsx
- analysis/16S/supplementary_table_ndvi_regression_16s.xlsx 0 additions, 0 deletionsanalysis/16S/supplementary_table_ndvi_regression_16s.xlsx
- analysis/FITS1/FITS1_Analysis_pipeline_v4-3.Rmd 22 additions, 11 deletionsanalysis/FITS1/FITS1_Analysis_pipeline_v4-3.Rmd
- analysis/FITS1/FITS1_Analysis_pipeline_v4-3.html 89 additions, 76 deletionsanalysis/FITS1/FITS1_Analysis_pipeline_v4-3.html
Source diff could not be displayed: it is too large. Options to address this: view the blob.
No preview for this file type
File deleted
No preview for this file type
171 KiB
File deleted
File deleted
File deleted
171 KiB
No preview for this file type
File deleted
No preview for this file type
File deleted
No preview for this file type
File deleted
No preview for this file type
No preview for this file type
This diff is collapsed.